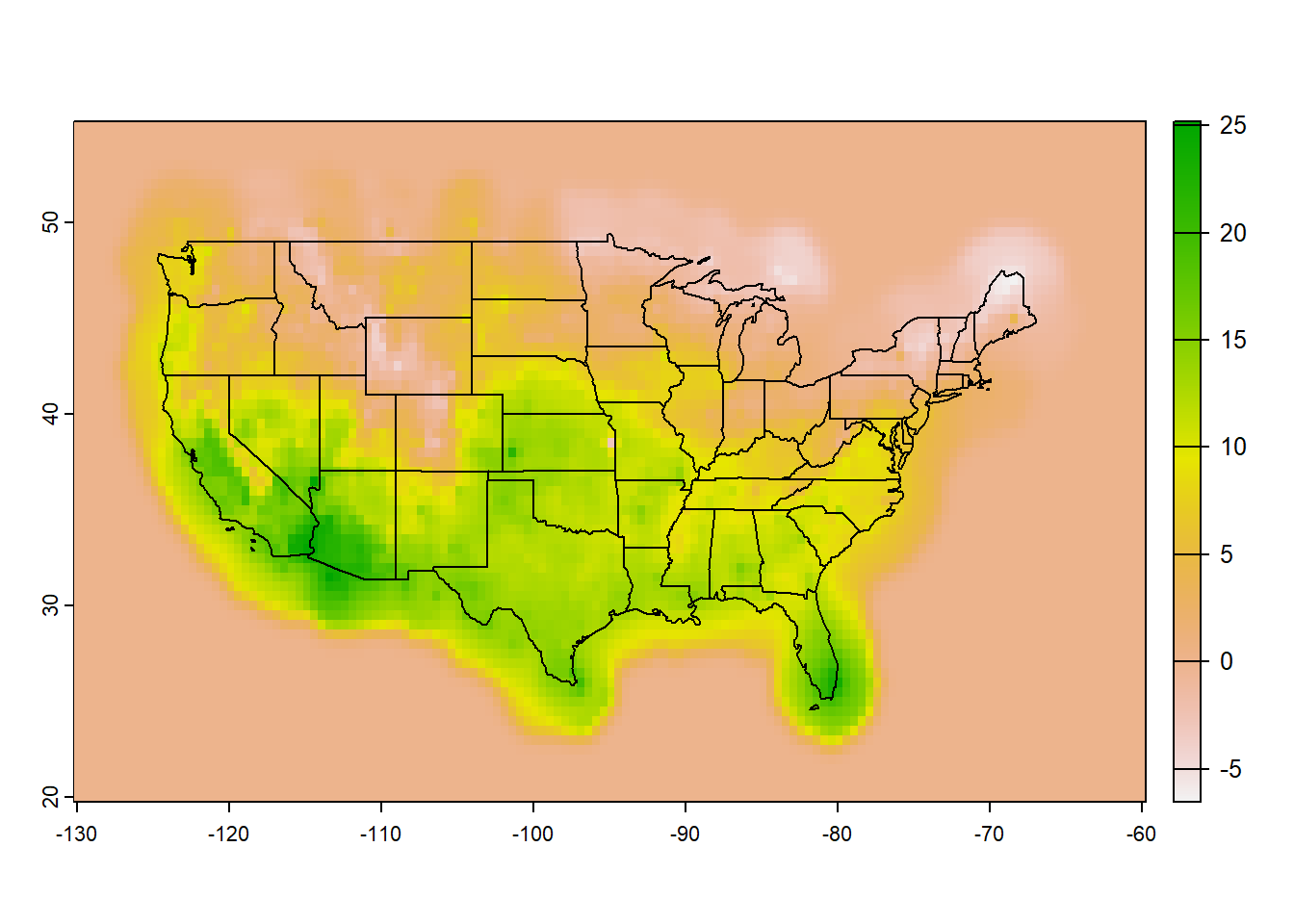
# Copied path of the raster
data_path <- "https://ftp.cpc.ncep.noaa.gov/GIS/GRADS_GIS/GeoTIFF/TEMP/us_tmax/us.tmax_nohads_ll_20240218_float.tif"
# Download the raster using download.file, assign the name tmax_20240218.tif to the downloaded
download.file(url = data_path,
method="curl",
destfile = "tmax_20240218.tif")
# Plot downloaded file
library(terra)
tempRas=rast("tmax_20240218.tif") # Import raster to the environment
usSHP=terra::vect(spData::us_states) # Shapefile for CONUS
plot(tempRas)
plot(usSHP, add=TRUE)6.1 Popular Climate Data Format
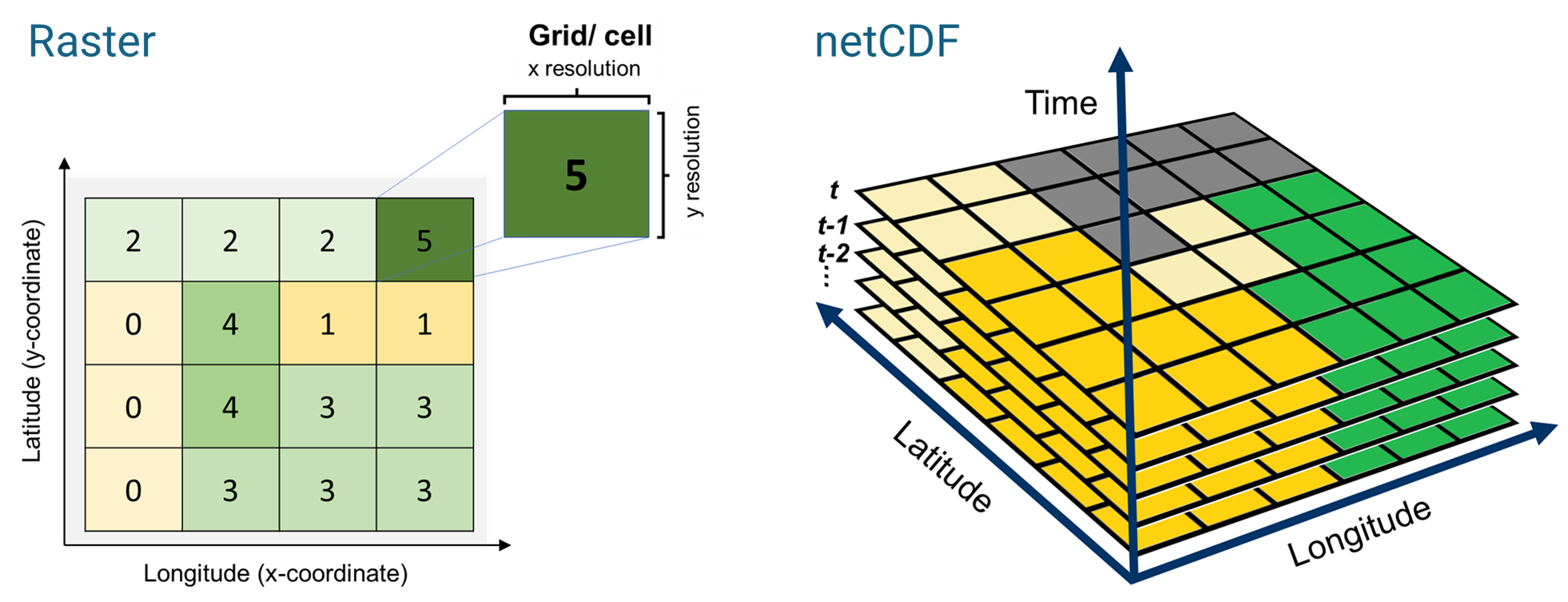
Raster and netCDF are two popular formats used for gridded climate data dissemination and archiving.
We are all too familiar with the raster format (pixelated, georeferenced data) from the previous chapters. NetCDF (Network Common Data Form), is a common data type for multi-layered, structured, gridded dataset. NetCDF is a machine-independent data format and is a community standard for sharing scientific data. A netCDF has certain features which makes it suitable for complex scientific data archiving and sharing, namely,
Self-Describing. A netCDF file includes information about the data it contains.
Portable. A netCDF file can be accessed by computers with different ways of storing integers, characters, and floating-point numbers.
Scalable. A small subset of a large dataset may be accessed efficiently.
Appendable. Data may be appended to a properly structured netCDF file without copying the dataset or redefining its structure.
Shareable. One writer and multiple readers may simultaneously access the same netCDF file.
Archivable. Access to all earlier forms of netCDF data will be supported by current and future versions of the software.
- From [Unidata | NetCDF (ucar.edu) https://www.unidata.ucar.edu/software/netcdf/]
Several open-source plaforms and agencies provide open access to a multitude of gridded land and climate datasets generated using satellites and land-surface/ climate models. In the following sections, we will familiarize ourselves with some of these resources.
6.2 Open-Data Platforms
6.2.1 Climate Data from NOAA Physical Sciences Lab
- Product overview / Data access/ Information: https://psl.noaa.gov/data/gridded/tables/daily.html

This website provides several land and climate variables such as: CPC Global Unified Gauge-Based Analysis of Daily Precipitation, CPC Global Temperature, NCEP/NCAR Reanalysis, Livneh daily CONUS near-surface gridded meteorological and derived hydrometeorological data.
6.2.2 DAYMET: Daily Gridded Weather and Climate Data for North America [1 km x 1 km]
Product overview: https://daymet.ornl.gov/
Data access: https://daac.ornl.gov/cgi-bin/dataset_lister.pl?p=32
Data information: Refer to the User Guide provided with each dataset.
Daymet provides long-term, continuous, gridded estimates of daily weather and climatology variables at 1 km grid resolution for North America. The dataset is available in several forms, including monthly and annual climate summaries, in addition to the daily and/or sub-daily climate forcings:
Daymet Version 4 Monthly Latency: Daily Surface Weather Data
Annual Climate Summaries on a 1-km Grid for North America, Version 4 R1
Monthly Climate Summaries on a 1-km Grid for North America, Version 4 R1
Station-Level Inputs and Cross-Validation for North America, Version 4 R1
Daily Surface Weather Data on a 1-km Grid for North America, Version 4 R1

6.2.3 CHIRPS Global Precipitation [~5 km x 5km]
Product overview: https://climatedataguide.ucar.edu/climate-data/chirps-climate-hazards-infrared-precipitation-station-data-version-2
Data access: https://data.chc.ucsb.edu/products/CHIRPS-2.0/
Data information: https://data.chc.ucsb.edu/products/CHIRPS-2.0/docs/README-CHIRPS.txt

6.2.4 GIMMS MODIS Global NDVI [~225 m x 225 m]
Product overview: https://ladsweb.modaps.eosdis.nasa.gov/missions-and-measurements/products/MOD13Q1
Data access: https://gimms.gsfc.nasa.gov/MODIS/
Data information: https://gimms.gsfc.nasa.gov/MODIS/README-global.txt

6.2.5 Climate Prediction Center
- Product overview / Data access/ Information: https://ftp.cpc.ncep.noaa.gov/GIS/

Includes several variables including (but not limited to):
Climate Prediction Center (CPC) Morphing Technique (MORPH) to form a global, high resolution precipitation analysis
Joint Agricultural Weather Facility (JAWF)
Grid Analysis and Display System (GRADS): Global precipitation monitoring and forecasts, Tmax, Tmin
Input variables for US Drought Monitor (USDM)
6.2.6 Soil Texture for CONUS [30 m x 30 m]
Probabilistic Remapping of SSURGO (POLARIS) is a database of 30-m probabilistic soil property maps over the Contiguous United States (CONUS) generated by removing artificial discontinuities in Soil Survey Geographic (SSURGO) database. using an artificial intelligence algorithm (Chaney et al. 2016, 2019). Estimates provided by POLARIS include soil texture, organic matter, pH, saturated hydraulic conductivity, Brooks-Corey and Van Genuchten water retention curve parameters, bulk density, and saturated water content for six profile depths, namely, 0-5 cm, 5-15 cm, 15-30 cm, 30-60 cm, 60-100 cm, 100-200 cm.
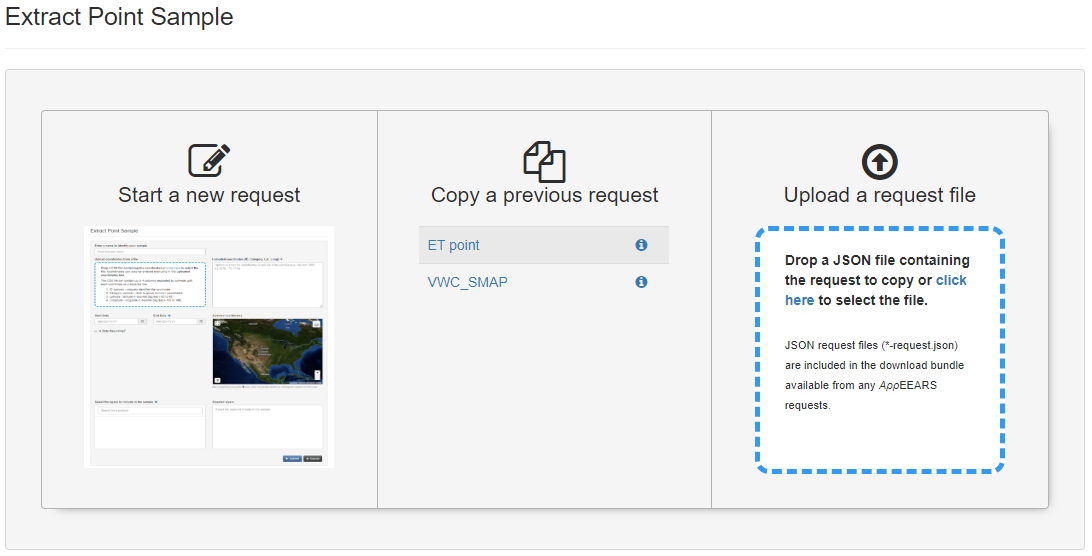
6.2.7 NASA AρρEEARS
- Product overview / Data access/ Information: https://appeears.earthdatacloud.nasa.gov/

Climate/ land data variables can be extracted for an area or a point using the interactive interface. Click on point samples/ area samples:
6.2.8 NASA Earth Data Search
- Product overview / Data access/ Information: https://search.earthdata.nasa.gov/search

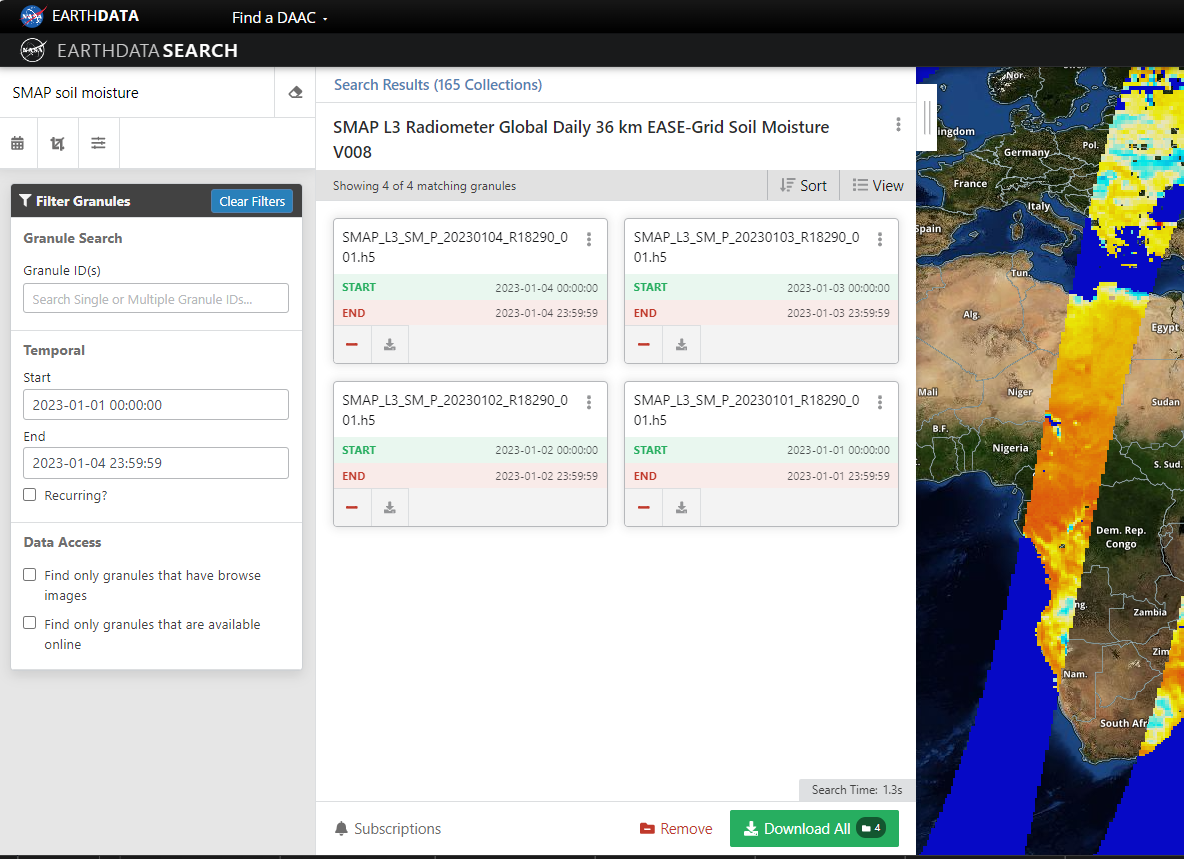
6.2.8.1 Bulk Download Order
NASA Earth Data provides customization options for bulk data download. Lets say we are interested in downloading global SMAP Level 3 soil moisture. We start by selecting the product, and specify start/ end date as needed.
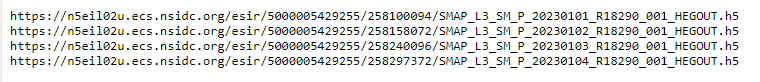
Copy and Paste these links in any internet download manager (my favorite in Chrono for Google Chrome), Select output location (typically an external hard drive) and let the download begin.
6.3 Programmatic Data Acquisition
In the HTTP, FTP or HTP links provided before, one can download a file by clicking on the individual hyperlink. Alternatively, we can use download.file function to download the file programmatically in R. This help us by opening the path to automate download and processing of multiple files with minimal supervision.
6.3.1 Downloading Raster files
Let us take an example of us_tmax data available at: https://ftp.cpc.ncep.noaa.gov/GIS/GRADS_GIS/GeoTIFF/TEMP/us_tmax/
Right-click on the raster file for 20240218, and copy the file path. We will then use this link to access the files programmatically using Client URL, or cURL - a utility for transferring data between systems. We will download the raster using download.file to local disk, and saved with a uder-defined name tmax_20240218.tif.
Copied link: https://ftp.cpc.ncep.noaa.gov/GIS/GRADS_GIS/GeoTIFF/TEMP/us_tmax/us.tmax_nohads_ll_20240218_float.tif
Now that we have the tmax_20240218 raster, let us extract the values for certain selected locations: s
# Import sample locations from contrasting hydroclimate
library(readxl)
loc= read_excel("./SampleData-master/location_points.xlsx")
print(loc)# A tibble: 3 × 4
Aridity State Longitude Latitude
<chr> <chr> <dbl> <dbl>
1 Humid Louisiana -92.7 34.3
2 Arid Nevada -116. 38.7
3 Semi-arid Kansas -99.8 38.8# Value of the lat & lon of the locations
latlon=loc[,3:4]
print(latlon)# A tibble: 3 × 2
Longitude Latitude
<dbl> <dbl>
1 -92.7 34.3
2 -116. 38.7
3 -99.8 38.8# Extract time series using "terra::extract"
loc_temp=terra::extract(tempRas,
latlon, #2-column matrix or data.frame with lat-long
method='bilinear') # Use bilinear interpolation (or ngb) option
print(loc_temp) ID tmax_20240218
1 1 12.58291
2 2 11.52682
3 3 13.41145# Add temperature attribute to the data frame as "temp"
loc$temp=loc_temp$tmax_20240218
print(loc)# A tibble: 3 × 5
Aridity State Longitude Latitude temp
<chr> <chr> <dbl> <dbl> <dbl>
1 Humid Louisiana -92.7 34.3 12.6
2 Arid Nevada -116. 38.7 11.5
3 Semi-arid Kansas -99.8 38.8 13.4# Export the modified data as CSV
write.csv(loc, "df_with_temp.csv")6.3.2 Raster Mosaic
Lets explore raster files for mean clay percentage for top 5 cm soil profile accessible through: http://hydrology.cee.duke.edu/POLARIS/PROPERTIES/v1.0/clay/mean/0_5/. The link provides rasters for 1x1 degree areal domain.
library(spData)
# Note the spatial extent of Louisiana State. We need corresponding raster files
ext(spData::us_states[spData::us_states$NAME=="Louisiana",]) # ext function gives extent of the rast/vect objectSpatExtent : -94.042964, -89.066617, 28.991623, 33.019219 (xmin, xmax, ymin, ymax)pth1="http://hydrology.cee.duke.edu/POLARIS/PROPERTIES/v1.0/clay/mean/0_5/lat3031_lon-91-90.tif"
# Download the raster using download.file
download.file(url = pth1,
method="curl",
destfile = "lat3031_lon-90-89.tif")
# Import the downloaded raster to workspace
r1=rast("lat3031_lon-90-89.tif")
# Fetch shapefile for Louisiana from spData package
LAvct=vect(spData::us_states[spData::us_states$NAME=="Louisiana",])
# Plot downloaded raster against the map of Louisiana. Note that the raster fits 1x1 grid
plot(LAvct, col="gray90")
plot(r1, range=c(0,100), add=TRUE)
grid()# Crop raster to smaller region of interest for easy visualization
#~~~ We will crop raster to a user-defined extent
r1crp=crop(r1, ext(c(-91,-90.8,30,30.2)))
# Explore a smaller subset of the dataset
#~~~ Can you identify the Mississipi flood-plain using the clay percentage?
library(mapview)
mapview(r1crp, # Raster to be plotted
at=c(0,10,20,30,40,50,60,75), # Legend breaks
map.types="Esri.WorldImagery", # Select background map
main="Clay % (0-5 cm profile)") # Plot titleWe note that the raster files in POLARIS are available for a 1x1 areal domain. For an analysis for a large spatial extent, multiple smaller rasters can be “stitched” together to generate a larger mosaic of rasters. We will use the terra::mosaic function to generate a mosaic of several smaller rasters of percentage clay content in 0-5 cm soil profile in Southeastern Louisiana. This function requires the user to specify the summary function (“sum”, “mean”, “median”, “min”, or “max”) to be applied on the overlapping pixels from 2 or more rasters. Let us download some more rasters from POLARIS and mosaic them together.
# Links to soil texture rasters
pth2="http://hydrology.cee.duke.edu/POLARIS/PROPERTIES/v1.0/clay/mean/0_5/lat2930_lon-91-90.tif"
pth3="http://hydrology.cee.duke.edu/POLARIS/PROPERTIES/v1.0/clay/mean/0_5/lat3031_lon-90-89.tif"
pth4="http://hydrology.cee.duke.edu/POLARIS/PROPERTIES/v1.0/clay/mean/0_5/lat3031_lon-92-91.tif"
# Download the raster using download.file function
download.file(url = pth2,
method="curl",
destfile = "r2.tif")
download.file(url = pth3,
method="curl",
destfile = "r3.tif")
download.file(url = pth4,
method="curl",
destfile = "r4.tif")
# Import downloaded files to workspace
r2=rast("r2.tif")
r3=rast("r3.tif")
r4=rast("r4.tif")
# DIY: Plot the downloaded raster against the map of Louisiana
# plot(LAvct)
# plot(r1, range=c(0,100), add=TRUE)
# plot(r2, range=c(0,100), add=TRUE, legend=FALSE)
# plot(r3, range=c(0,100), add=TRUE, legend=FALSE)
# plot(r4, range=c(0,100), add=TRUE, legend=FALSE)
# Raster mosaic
r_mos=mosaic(r1,r2,r3,r4, fun="mean")
# Plot raster mossaic
plot(LAvct, main="Clay % [0-5 cm depth]",axes=FALSE, col="gray90")
plot(r_mos, range=c(0,100), add=TRUE)
plot(LAvct, add=TRUE)
grid()For easy post processing access, its a good practice to save the processed dataset on disc as either a raster or netCDF.
# Export raster to disc
terra::writeRaster(r_mos, "clayPct.tif",
overwrite=TRUE) # Overwrite if the file already exists?
# Or export as netCDF
terra::writeCDF(r_mos, "clayPct.nc",
overwrite=TRUE) # Overwrite if the file already exists?
# Optional: Add more information to the exported netCDF
# terra::writeCDF(r_mos, # SpatRaster to export
# "clayPct.nc", # Output filename
# varname="clayPCT", # Short name of the variable
# unit="%", # Variable units
# longname="Clay Percentage, 0-5 cm, POLARIS", # Long name of the variable
# zname='[-]') # Z-var name (None, since we export a single layer)6.3.3 Downloading netCDF
We will now download a netCDF of global daily precipitation for the year 2023 from CHIRPS, accessible through the link: https://data.chc.ucsb.edu/products/CHIRPS-2.0/global_daily/netcdf/p05/chirps-v2.0.2023.days_p05.nc
# Copied path of the raster
data_path <- "https://data.chc.ucsb.edu/products/CHIRPS-2.0/global_daily/netcdf/p05/chirps-v2.0.2023.days_p05.nc"
# Download the raster using download.file, assign the name "daily_pcp_2023.nc" to the downloaded
if (file.exists("daily_pcp_2023.nc")==FALSE){
download.file(url = data_path,
method="curl",
destfile = "daily_pcp_2023.nc")
}
# Plot downloaded file
library(terra)
pcp=rast("daily_pcp_2023.nc") # Import raster to the environment
print(pcp) # Notice the attributes (esp. nlyr, i.e. number of layers, unit and time)class : SpatRaster
dimensions : 2000, 7200, 365 (nrow, ncol, nlyr)
resolution : 0.05, 0.05 (x, y)
extent : -180, 180, -50, 50 (xmin, xmax, ymin, ymax)
coord. ref. : +proj=longlat +datum=WGS84 +no_defs
source : daily_pcp_2023.nc
varname : precip (Climate Hazards group InfraRed Precipitation with Stations)
names : precip_1, precip_2, precip_3, precip_4, precip_5, precip_6, ...
unit : mm/day, mm/day, mm/day, mm/day, mm/day, mm/day, ...
time (days) : 2023-01-01 to 2023-12-31 head(time(pcp)) # time variable in the netCDF indicating corresponding time of acquisition[1] "2023-01-01" "2023-01-02" "2023-01-03" "2023-01-04" "2023-01-05"
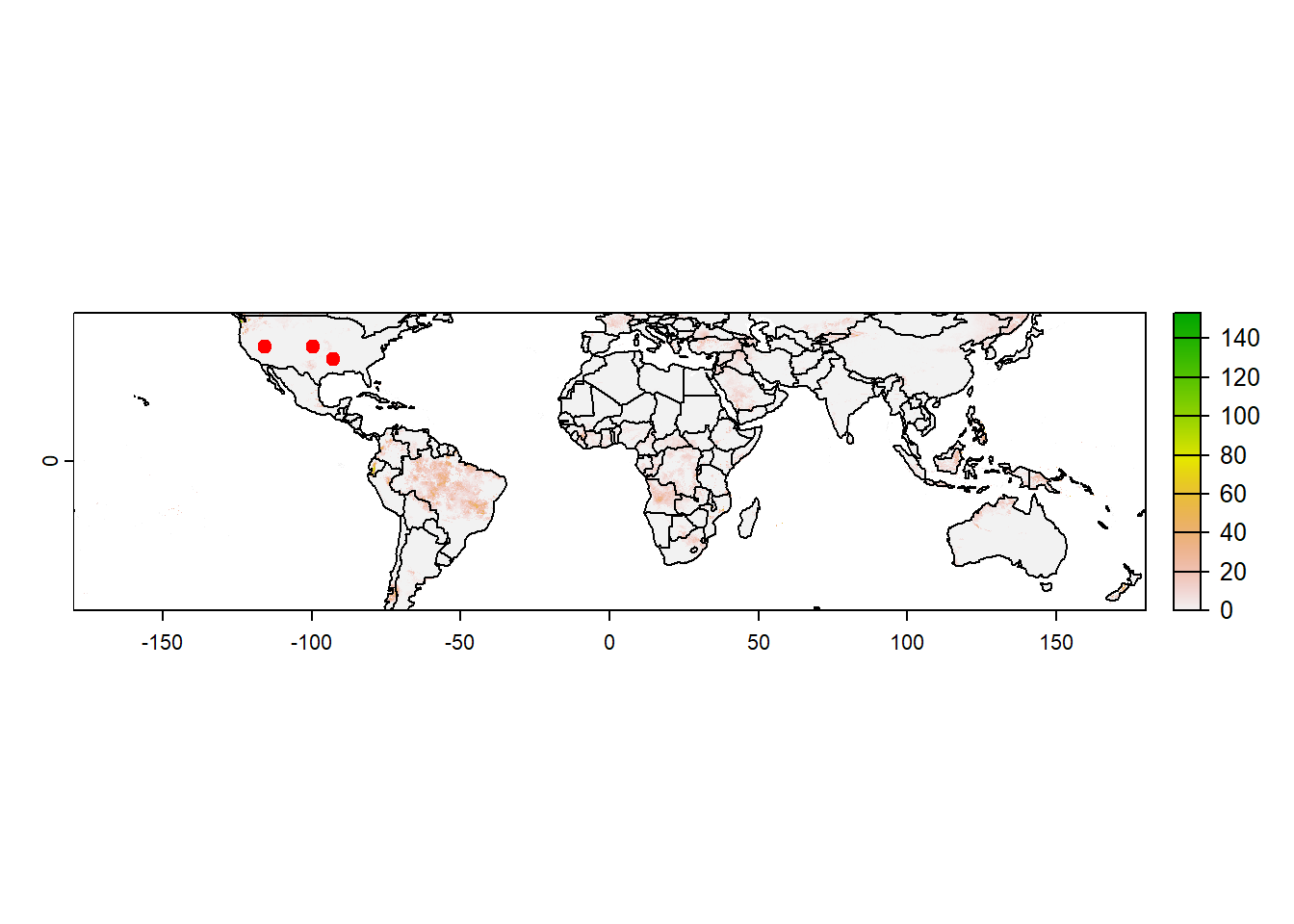
[6] "2023-01-06"worldSHP=terra::vect(spData::world) # Shapefile for CONUS
# Plot data for a specific layer
plot(pcp[[100]]) # Same as pcp[[which(time(pcp)=="2023-04-10")]]
plot(worldSHP, add=TRUE)
points(latlon, pch=19, col="red")